How to downsample a tractogram
By default, our pipeline does not save the tractograms or compress them after connectome calculations (-SC). The main reason is storage and computational time, which is a significant issue even when compression is used (for 10 million streamlines, uncompressed/compressed tractograms are ~14G/3G per case). When running the pipeline, it is possible to keep tractograms (optional argument -keep_tck), but it’s up to the final user to compress them.
Below you can check the estimated sizes and computation times of different tractograms.
Streamlines |
Original size MB |
Reduced size MB |
Computing time (seconds) |
10,000 |
19 |
4 |
2.86 |
100,000 |
152 |
32 |
21.05 |
1,000,000 |
1,428 |
303 |
479.52 |
10,000,000 |
1,4262 |
3,019 |
28920.14 |
Our example is based on the tutorial provided by DIPY called Streamline length and size reduction, but applied to micapipe’s outputs.
After a tractogram was saved (using the flags -SC -keep_tck), it could be downsampled using DIPY 1.5.0 and nibabel 3.2.1 tools.
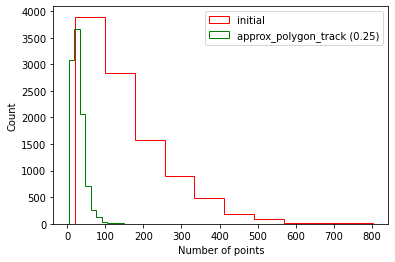
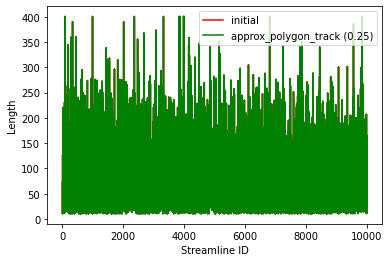
We use the function approx_polygon_track which reduces the number of points on each streamline without changing the length or shape, thus is an ideal methods for lossy compression of streamlines.
The following figures show how this procedure reduces the number of points without changing the length, therefore reducing the file size (we used a 10,000 streamlines tractogram for the figures).
Code example in python
Bellow you can find an example of downsampling a tractogram. The code is mean to run from the derivatives/micapipe directory:
1# Set enviroment
2import nibabel as nib
3from dipy.io.streamline import load_tractogram, save_tractogram
4from dipy.tracking.distances import approx_polygon_track
5
6# Function to downsample a tractogram
7def compress_tck(file_name):
8
9 # Load reference anatomy
10 reference_anatomy=nib.load(subject + '/dwi/' + subject + '_ses-01_space-dwi_desc-b0.nii.gz')
11
12 # load tractogram
13 bundle = load_tractogram(file_name, reference_anatomy, bbox_valid_check=False)
14 print('This bundle has %d streamlines' % len(bundle))
15
16 #tractogram size reduction
17 bundle_downsampled = [approx_polygon_track(s, 0.25) for s in bundle.streamlines]
18
19 # Replace the tractogram with the downsample
20 bundle.streamlines = bundle_downsampled
21
22 # Save new TCK file
23 save_tractogram(bundle, file_name.replace('.tck','_downsampled.tck'))
24 print('Downsampled tractogram was saved')
25
26# Compress a tractogram
27subject = 'sub-01'
28tck = subject + '/dwi/' + subject + '_space-dwi_desc-iFOD1-10M_tractography.tck'
29compress_tck(tck)