Surface visualization
Table of Contents
This section describes the whereabouts of the main surfaces generated by the pipeline, and how you can visualized them using python or R.
This example will use the subject HC001 and session 01 from the MICs dataset, and all paths will be relative to the subject directory or out/micapipe/sub-HC001_ses01/
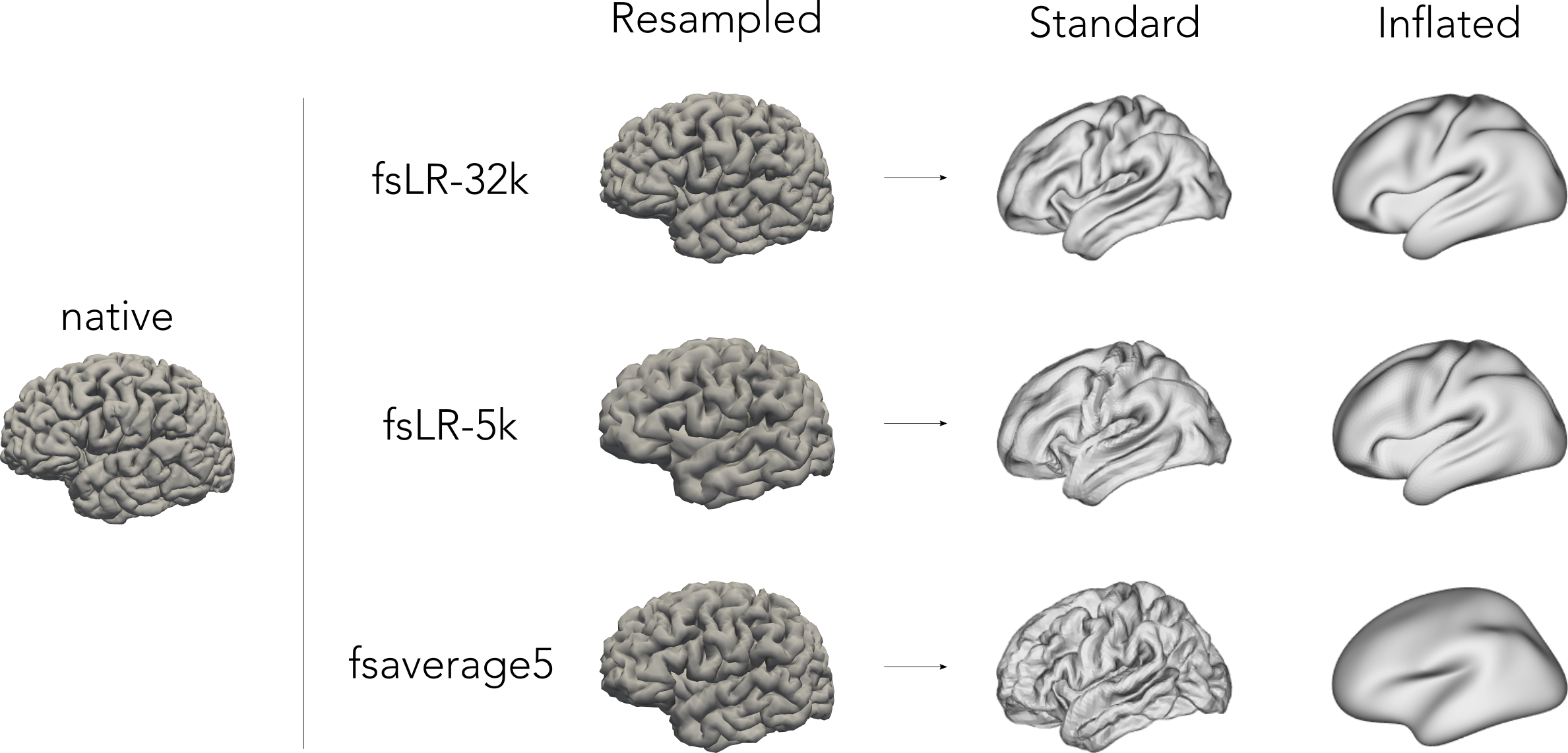
In the following examples, we’ll load and visualize single subject surfaces. Surfaces are distributed across different directories, namely:
sub-HC001/
└── ses-01
├── surf # fsnative, fsaverage5, fsLR-32k, fsLR-5k surfaces**
└── maps # Thickness, curvature and quantitative maps**
Each native surface parcellation is found inside the subject’s freesurfer directory and contains the string mics.annot:
freesurfer/
└── sub-HC001_ses-01
└── label
├── lh.schaefer-400_**mics.annot**
└── rh.schaefer-400_**mics.annot**
Setting the environment
This example uses the packages the python packages os, matplotlib, numpy, nibabel, matplotlib and brainspace.
If you are interested in plotting surfaces with BrainSpace, check the corresponding documentation!!
R libraries are RColorBrewer, viridis, fsbrain, freesurferformats and rgl.
For further information about managing and visualizing surfaces with R, check the fsbrain vignettes, fsbrain github repository, and
freesurferformats
The first step in both languages is to set the environment.
# Set the environment
import os
import glob
import numpy as np
import nibabel as nib
import seaborn as sns
from brainspace.plotting import plot_hemispheres
from brainspace.mesh.mesh_io import read_surface
from brainspace.datasets import load_conte69
# Set the working directory to the 'out' directory
out='/data_/mica3/BIDS_MICs/derivatives' # <<<<<<<<<<<< CHANGE THIS PATH
os.chdir(out)
# This variable will be different for each subject
sub='sub-HC001'
ses='ses-01'
subjectID=f'{sub}_{ses}' # <<<<<<<<<<<< CHANGE THIS SUBJECT's ID
subjectDir=f'micapipe_v0.2.0/{sub}/{ses}' # <<<<<<<<<<<< CHANGE THIS SUBJECT's DIRECTORY
# Set paths and variables
dir_FS = 'freesurfer/' + subjectID
dir_surf = subjectDir + '/surf/'
dir_maps = subjectDir + '/maps/'
# Path to MICAPIPE
micapipe=os.popen("echo $MICAPIPE").read()[:-1]
# Set the environment 'R 3.6.3'
require('RColorBrewer') # version 1.1-2
require('viridis') # version 0.5.1
require('fsbrain') # version 0.4.2
require('freesurferformats') # version 0.1.14
require('rgl') # version 0.1.54
# Set the working directory to the out directory
setwd("~/tmp/micaConn/micapipe_tutorials") # <<<<<<<<<<<< CHANGE THIS PATH
# This variable will be different for each subject
subjectID <- 'sub-HC001_ses-01' # <<<<<<<<<<<< CHANGE THIS SUBJECT's ID
subjectDir <- 'micapipe/sub-HC001/ses-01' # <<<<<<<<<<<< CHANGE THIS SUBJECT's DIRECTORY
# Here we define the atlas
atlas <- 'schaefer-400' # <<<<<<<<<<<< CHANGE THIS ATLAS
# Set paths and variables
dir_surf <- paste0(subjectDir, '/surf/')
dir_maps <- paste0(subjectDir, '/maps/')
Load the surfaces
# Load native pial surface
pial_lh = read_surface(dir_FS+'/surf/lh.pial', itype='fs')
pial_rh = read_surface(dir_FS+'/surf/rh.pial', itype='fs')
# Load native white matter surface
wm_lh = read_surface(dir_FS+'/surf/lh.white', itype='fs')
wm_rh = read_surface(dir_FS+'/surf/rh.white', itype='fs')
# Load native inflated surface
inf_lh = read_surface(dir_FS+'/surf/lh.inflated', itype='fs')
inf_rh = read_surface(dir_FS+'/surf/rh.inflated', itype='fs')
# Load fsaverage5
fs5_lh = read_surface('freesurfer/fsaverage5/surf/lh.pial', itype='fs')
fs5_rh = read_surface('freesurfer/fsaverage5/surf/rh.pial', itype='fs')
# Load fsaverage5 inflated
fs5_inf_lh = read_surface('freesurfer/fsaverage5/surf/lh.inflated', itype='fs')
fs5_inf_rh = read_surface('freesurfer/fsaverage5/surf/rh.inflated', itype='fs')
# Load fsLR 32k
f32k_lh = read_surface(micapipe + '/surfaces/fsLR-32k.L.surf.gii', itype='gii')
f32k_rh = read_surface(micapipe + '/surfaces/fsLR-32k.R.surf.gii', itype='gii')
# Load fsLR 32k inflated
f32k_inf_lh = read_surface(micapipe + '/surfaces/fsLR-32k.L.inflated.surf.gii', itype='gii')
f32k_inf_rh = read_surface(micapipe + '/surfaces/fsLR-32k.R.inflated.surf.gii', itype='gii')
# Load Load fsLR 5k
f5k_lh = read_surface(micapipe + '/surfaces/fsLR-5k.L.surf.gii', itype='gii')
f5k_rh = read_surface(micapipe + '/surfaces/fsLR-5k.R.surf.gii', itype='gii')
# Load fsLR 5k inflated
f5k_inf_lh = read_surface(micapipe + '/surfaces/fsLR-5k.L.inflated.surf.gii', itype='gii')
f5k_inf_rh = read_surface(micapipe + '/surfaces/fsLR-5k.R.inflated.surf.gii', itype='gii')
# Helper function
plot_surface <-function(brainMesh, legend='', view_angles=c('sd_lateral_lh', 'sd_medial_lh', 'sd_medial_rh', 'sd_lateral_rh'), img_only=FALSE) {
try(img <- vis.export.from.coloredmeshes(brainMesh, colorbar_legend = legend, grid_like = FALSE, view_angles = view_angles, img_only = img_only, horizontal=TRUE))
while (rgl.cur() > 0) { rgl.close() }; file.remove(list.files(path = getwd(), pattern = 'fsbrain'))
return(img)
}
Morphology
Two surface based morphological features are plotted here: cortical thickness and curvature. Both measurements are generates in three main surfaces, native, fsaverage5, fsLR-32k and fsLR-5k.
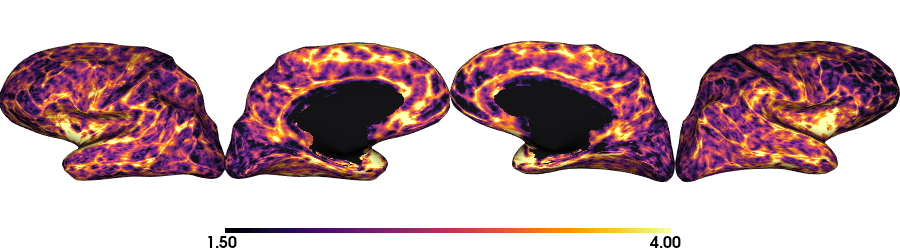
Thickness: Inflated native surface
# Load data
th_lh = dir_maps + subjectID + '_hemi-L_surf-fsnative_label-thickness.func.gii'
th_rh = dir_maps + subjectID + '_hemi-R_surf-fsnative_label-thickness.func.gii'
th_nat = np.hstack(np.concatenate((nib.load(th_lh).darrays[0].data,
nib.load(th_rh).darrays[0].data), axis=0))
# Plot the surface
plot_hemispheres(inf_lh, inf_rh, array_name=th_nat, size=(900, 250), color_bar='bottom', zoom=1.25, embed_nb=True, interactive=False, share='both',
nan_color=(0, 0, 0, 1), color_range=(1.5, 4), cmap="inferno", transparent_bg=False)
# Set the path to the surface
th.lh <- paste0(dir_maps, subjectID, "_hemi-L_surf-fsnative_label-thickness.func.gii")
th.rh <- paste0(dir_maps, subjectID, "_hemi-R_surf-fsnative_label-thickness.func.gii")
# Plot the surface
th_nat <- vis.data.on.subject('freesurfer/', subjectID, morph_data_lh=th.lh, morph_data_rh=th.rh, surface="inflated", draw_colorbar = TRUE,
views=NULL, rglactions = list('trans_fun'=limit_fun(1.5, 4), 'no_vis'=T), makecmap_options = list('colFn'=inferno))
plot_surface(th_nat, 'Thickness [mm]')
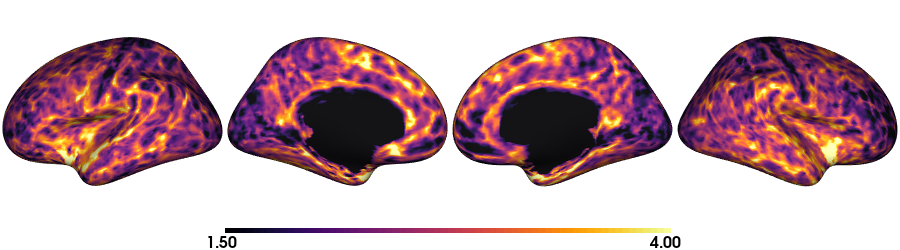
Thickness: fsaverage5
# Load data
th_lh_fs5 = dir_maps + subjectID + '_hemi-L_surf-fsaverage5_label-thickness.func.gii'
th_rh_fs5 = dir_maps + subjectID + '_hemi-R_surf-fsaverage5_label-thickness.func.gii'
th_fs5 = np.hstack(np.concatenate((nib.load(th_lh_fs5).darrays[0].data,
nib.load(th_rh_fs5).darrays[0].data), axis=0))
# Plot the surface
plot_hemispheres(fs5_inf_lh, fs5_inf_rh, array_name=th_fs5, size=(900, 250), color_bar='bottom', zoom=1.25, embed_nb=True, interactive=False, share='both',
nan_color=(0, 0, 0, 1), color_range=(1.5, 4), cmap="inferno", transparent_bg=False)
# Set the path to the surface
th.lh.fs5 <- paste0(dir_maps, subjectID, "_hemi-L_surf-fsaverage5_label-thickness.func.gii")
th.rh.fs5 <- paste0(dir_maps, subjectID, "_hemi-R_surf-fsaverage5_label-thickness.func.gii")
# Plot the surface
th_fs5 <- vis.data.on.subject('freesurfer/', 'fsaverage5', morph_data_lh=th.lh.fs5, morph_data_rh=th.rh.fs5, surface="inflated", draw_colorbar = TRUE,
views=NULL, rglactions = list('trans_fun'=limit_fun(1.5, 4), 'no_vis'=T), makecmap_options = list('colFn'=inferno))
plot_surface(th_fs5, 'Thickness [mm]')
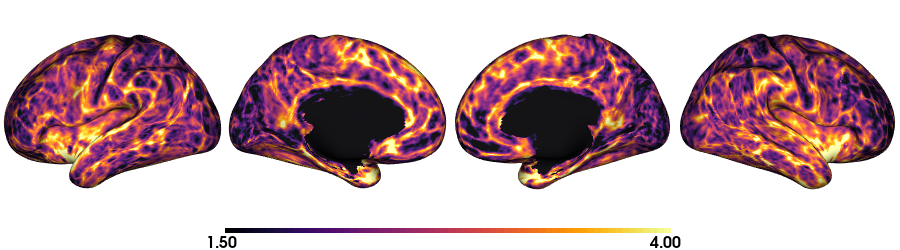
Thickness: fsLR-32k
# Load the data
th_lh_fsLR32k = dir_maps + subjectID + '_hemi-L_surf-fsLR-32k_label-thickness.func.gii'
th_rh_fsLR32k = dir_maps + subjectID + '_hemi-R_surf-fsLR-32k_label-thickness.func.gii'
th_fsLR32k = np.hstack(np.concatenate((nib.load(th_lh_fsLR32k).darrays[0].data,
nib.load(th_rh_fsLR32k).darrays[0].data), axis=0))
# Plot the surface
plot_hemispheres(f32k_inf_lh, f32k_inf_rh, array_name=th_fsLR32k, size=(900, 250), color_bar='bottom', zoom=1.25, embed_nb=True, interactive=False, share='both',
nan_color=(0, 0, 0, 1), color_range=(1.5, 4), cmap="inferno", transparent_bg=False)
# Set the path to the surface
th.lh.f32k <- paste0(dir_maps, subjectID, '_hemi-L_surf-fsLR-32k_label-thickness.func.gii')
th.rh.f32k <- paste0(dir_maps, subjectID, '_hemi-R_surf-fsLR-32k_label-thickness.func.gii')
# Plot the surface
th_f32k <- vis.data.on.subject('freesurfer/', 'fsLR-32k', morph_data_lh=th.lh.f32k, morph_data_rh=th.rh.f32k, surface='fsLR-32k.gii', draw_colorbar = TRUE,
views=NULL, rglactions = list('trans_fun'=limit_fun(1.5, 4), 'no_vis'=T), makecmap_options = list('colFn'=inferno))
plot_surface(th_f32k, 'Thickness [mm]')
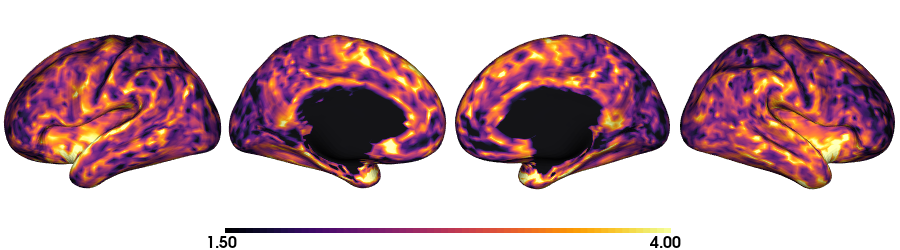
Thickness: fsLR-5k
# Load the data
th_lh_fsLR5k = dir_maps + subjectID + '_hemi-L_surf-fsLR-5k_label-thickness.func.gii'
th_rh_fsLR5k = dir_maps + subjectID + '_hemi-R_surf-fsLR-5k_label-thickness.func.gii'
th_fsLR5k = np.hstack(np.concatenate((nib.load(th_lh_fsLR5k).darrays[0].data,
nib.load(th_rh_fsLR5k).darrays[0].data), axis=0))
# Plot the surface
plot_hemispheres(f5k_inf_lh, f5k_inf_rh, array_name=th_fsLR5k, size=(900, 250), color_bar='bottom', zoom=1.25, embed_nb=True, interactive=False, share='both',
nan_color=(0, 0, 0, 1), color_range=(1.5, 4), cmap="inferno", transparent_bg=False)
# Set the path to the surface
th.lh.f5k <- paste0(dir_maps, subjectID, '_hemi-L_surf-fsLR-5k_label-thickness.func.gii')
th.rh.f5k <- paste0(dir_maps, subjectID, '_hemi-R_surf-fsLR-5k_label-thickness.func.gii')
# Plot the surface
th_f5k <- vis.data.on.subject('freesurfer/', 'fsLR-5k', morph_data_lh=th.lh.f5k, morph_data_rh=th.rh.f5k, surface='fsLR-5k.gii', draw_colorbar = TRUE,
views=NULL, rglactions = list('trans_fun'=limit_fun(1.5, 4), 'no_vis'=T), makecmap_options = list('colFn'=inferno))
plot_surface(th_f5k, 'Thickness [mm]')
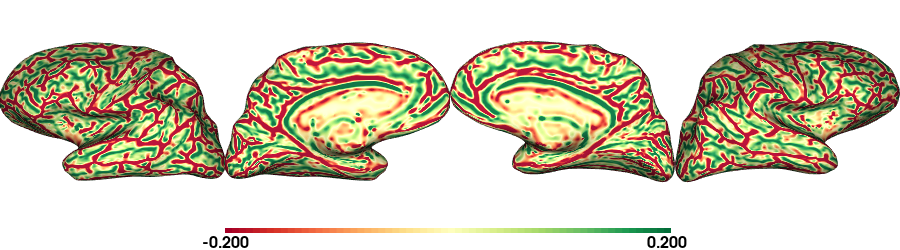
Curvature: Native inflated surface
# Load the data
cv_lh = dir_maps + subjectID + '_hemi-L_surf-fsnative_label-curv.func.gii'
cv_rh = dir_maps + subjectID + '_hemi-R_surf-fsnative_label-curv.func.gii'
cv = np.hstack(np.concatenate((nib.load(cv_lh).darrays[0].data,
nib.load(cv_rh).darrays[0].data), axis=0))
# Plot the surface
plot_hemispheres(inf_lh, inf_rh, array_name=cv, size=(900, 250), color_bar='bottom', zoom=1.25, embed_nb=True, interactive=False, share='both',
nan_color=(0, 0, 0, 1), color_range=(-0.2, 0.2), cmap='RdYlGn', transparent_bg=False)
# Colormap
RdYlGn <- colorRampPalette(brewer.pal(11,"RdYlGn"))
#### Curvature: Native surface
# Set the path to the surface
cv.lh <- paste0(dir_maps, subjectID, "_space-fsnative_desc-lh_curvature.mgh")
cv.rh <- paste0(dir_maps, subjectID, "_space-fsnative_desc-rh_curvature.mgh")
# Plot the surface
cv_nat <- vis.data.on.subject('freesurfer/', subjectID, morph_data_lh=cv.lh, morph_data_rh=cv.rh, surface="inflated", draw_colorbar = TRUE,
views=NULL, rglactions = list('trans_fun'=limit_fun(-0.2, 0.2), 'no_vis'=T), makecmap_options = list('colFn'=RdYlGn))
plot_surface(cv_nat, 'Curvature [1/mm]')
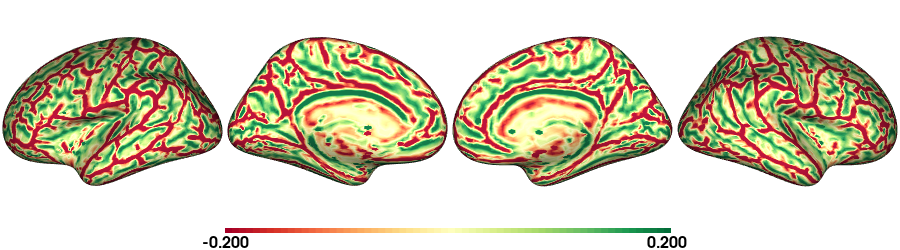
Curvature: fsaverage5
# Load the data
cv_lh_fs5 = dir_maps + subjectID + '_hemi-L_surf-fsaverage5_label-curv.func.gii'
cv_rh_fs5 = dir_maps + subjectID + '_hemi-R_surf-fsaverage5_label-curv.func.gii'
cv_fs5 = np.hstack(np.concatenate((nib.load(cv_lh_fs5).darrays[0].data,
nib.load(cv_rh_fs5).darrays[0].data), axis=0))
# Plot the surface
plot_hemispheres(fs5_inf_lh, fs5_inf_rh, array_name=cv_fs5, size=(900, 250), color_bar='bottom', zoom=1.25, embed_nb=True, interactive=False, share='both',
nan_color=(0, 0, 0, 1), color_range=(-0.2, 0.2), cmap='RdYlGn', transparent_bg=False)
# Set the path to the surface
cv.lh.fs5 <- paste0(dir_maps, subjectID, "_space-fsaverage5_desc-lh_curvature.mgh")
cv.rh.fs5 <- paste0(dir_maps, subjectID, "_space-fsaverage5_desc-rh_curvature.mgh")
# Plot the surface
cv_fs5 <- vis.data.on.subject('freesurfer/', 'fsaverage5', morph_data_lh=cv.lh.fs5, morph_data_rh=cv.rh.fs5, surface="inflated", draw_colorbar = TRUE,
views=NULL, rglactions = list('trans_fun'=limit_fun(-0.2, 0.2), 'no_vis'=T), makecmap_options = list('colFn'=RdYlGn))
plot_surface(cv_fs5, 'Curvature [1/mm]')
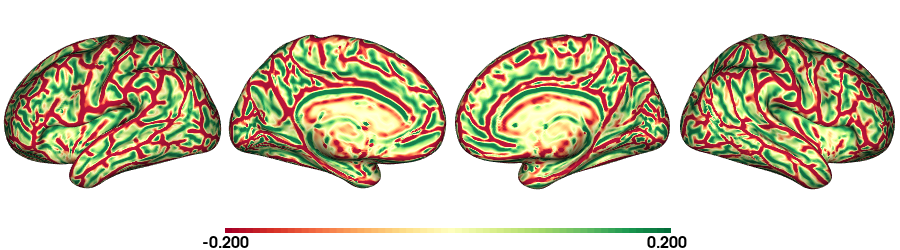
Curvature: fsLR-32k
# Load the data
cv_lh_fsLR32k = dir_maps + subjectID + '_hemi-L_surf-fsLR-32k_label-curv.func.gii'
cv_rh_fsLR32k = dir_maps + subjectID + '_hemi-R_surf-fsLR-32k_label-curv.func.gii'
cv_fsLR32k = np.hstack(np.concatenate((nib.load(cv_lh_fsLR32k).darrays[0].data,
nib.load(cv_rh_fsLR32k).darrays[0].data), axis=0))
# Plot the surface
plot_hemispheres(f32k_inf_lh, f32k_inf_rh, array_name=cv_fsLR32k, size=(900, 250), color_bar='bottom', zoom=1.25, embed_nb=True, interactive=False, share='both',
nan_color=(0, 0, 0, 1), color_range=(-0.2, 0.2), cmap='RdYlGn', transparent_bg=False)
# Set the path to the surface
cv.lh.f32k <- paste0(dir_maps, subjectID, '_space-fsLR-32k-32k_desc-lh_curvature.mgh')
cv.rh.f32k <- paste0(dir_maps, subjectID, '_space-fsLR-32k-32k_desc-rh_curvature.mgh')
# Plot the surface
cv_f32k <- vis.data.on.subject('freesurfer', 'fsLR-32k', morph_data_lh=cv.lh.f32k, morph_data_rh=cv.rh.f32k, surface='fsLR-32k.gii', draw_colorbar = TRUE,
views=NULL, rglactions = list('trans_fun'=limit_fun(-0.2, 0.2), 'no_vis'=T), makecmap_options = list('colFn'=RdYlGn))
plot_surface(cv_f32k, 'Curvature [1/mm]')
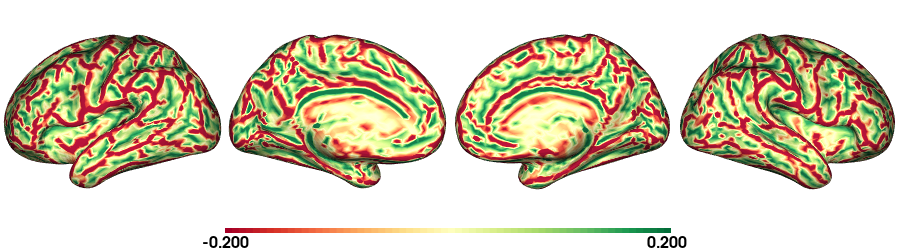
Curvature: fsLR-5k
# Load the data
cv_lh_fsLR5k = dir_maps + subjectID + '_hemi-L_surf-fsLR-5k_label-curv.func.gii'
cv_rh_fsLR5k = dir_maps + subjectID + '_hemi-R_surf-fsLR-5k_label-curv.func.gii'
cv_fsLR5k = np.hstack(np.concatenate((nib.load(cv_lh_fsLR5k).darrays[0].data,
nib.load(cv_rh_fsLR5k).darrays[0].data), axis=0))
# Plot the surface
plot_hemispheres(f5k_inf_lh, f5k_inf_rh, array_name=cv_fsLR5k, size=(900, 250), color_bar='bottom', zoom=1.25, embed_nb=True, interactive=False, share='both',
nan_color=(0, 0, 0, 1), color_range=(-0.2, 0.2), cmap='RdYlGn', transparent_bg=False)
# Set the path to the surface
cv.lh.f5k <- paste0(dir_maps, subjectID, '_space-fsLR-5k-5k_desc-lh_curvature.mgh')
cv.rh.f5k <- paste0(dir_maps, subjectID, '_space-fsLR-5k-5k_desc-rh_curvature.mgh')
# Plot the surface
cv_f5k <- vis.data.on.subject('freesurfer', 'fsLR-5k', morph_data_lh=cv.lh.f5k, morph_data_rh=cv.rh.f5k, surface='fsLR-5k.gii', draw_colorbar = TRUE,
views=NULL, rglactions = list('trans_fun'=limit_fun(-0.2, 0.2), 'no_vis'=T), makecmap_options = list('colFn'=RdYlGn))
plot_surface(cv_f5k, 'Curvature [1/mm]')
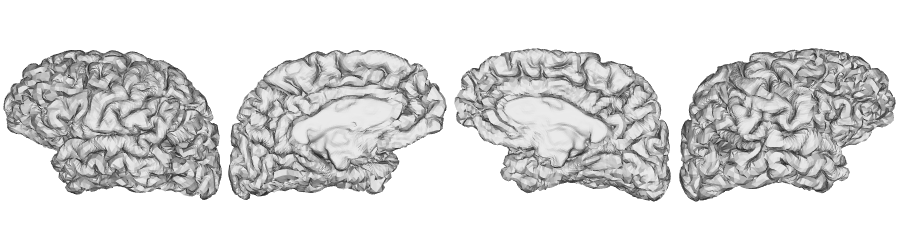
fsLR-32k
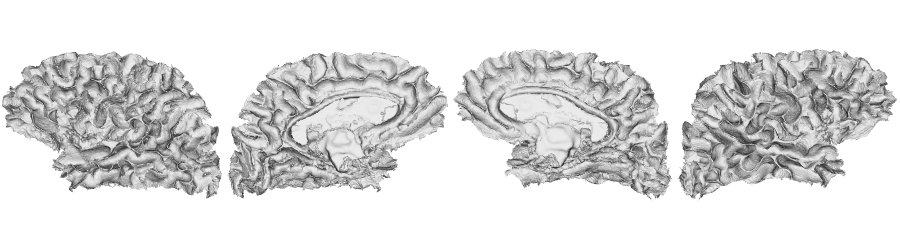
fsLR-32k: Pial surface
# Native conte69 pial surface
fsLR32k_pial_lh = read_surface(dir_surf+subjectID+'_hemi-L_space-nativepro_surf-fsLR-32k_label-pial.surf.gii', itype='gii')
fsLR32k_pial_rh = read_surface(dir_surf+subjectID+'_hemi-R_space-nativepro_surf-fsLR-32k_label-pial.surf.gii', itype='gii')
# Plot the surface
plot_hemispheres(fsLR32k_pial_lh, fsLR32k_pial_rh, size=(900, 250), zoom=1.25, embed_nb=True, interactive=False, share='both',
nan_color=(0, 0, 0, 1), color_range=(1.5, 4), cmap='Greys', transparent_bg=False)
# Colormap
grays <- colorRampPalette(c('gray65', 'gray65', 'gray65'))
# Set the path to the surface
f32k.pial.lh <- read.fs.surface(filepath = paste0(dir_surf, subjectID,'_space-fsLR-32k-32k_desc-lh_pial.surf.gii') )
f32k.pial.rh <- read.fs.surface(filepath = paste0(dir_surf, subjectID,'_space-fsLR-32k-32k_desc-rh_pial.surf.gii') )
# Plot the surface
cml = coloredmesh.from.preloaded.data(f32k.pial.lh, morph_data = rep(0, nrow(f32k.pial.lh$vertices)), makecmap_options = list('colFn'=grays) )
cmr = coloredmesh.from.preloaded.data(f32k.pial.rh, morph_data = rep(0, nrow(f32k.pial.rh$vertices)), makecmap_options = list('colFn'=grays) )
brainviews(views = 't4', coloredmeshes=list('lh'=cml, 'rh'=cmr), draw_colorbar = FALSE,
rglactions = list('trans_fun'=limit_fun(-1, 1), 'no_vis'=F))
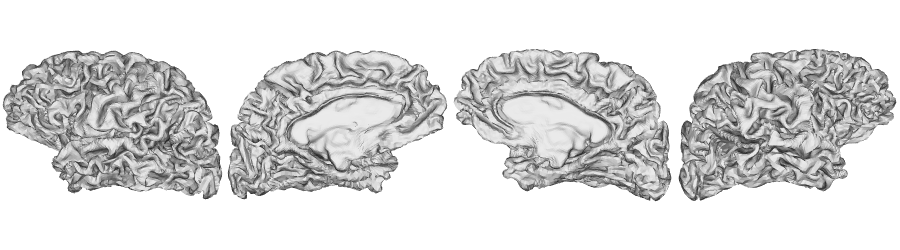
fsLR-32k: Middle surface
# Native fsLR-32k midsurface
fsLR32k_mid_lh = read_surface(dir_surf+subjectID+'_hemi-L_space-nativepro_surf-fsLR-32k_label-midthickness.surf.gii', itype='gii')
fsLR32k_mid_rh = read_surface(dir_surf+subjectID+'_hemi-R_space-nativepro_surf-fsLR-32k_label-midthickness.surf.gii', itype='gii')
# Plot the surface
plot_hemispheres(fsLR32k_mid_lh, fsLR32k_mid_rh, size=(900, 250), zoom=1.25, embed_nb=True, interactive=False, share='both',
nan_color=(0, 0, 0, 1), color_range=(-1,1), cmap='Greys', transparent_bg=False)
# Set the path to the surface
f32k.mid.lh <- read.fs.surface(filepath = paste0(dir_surf, subjectID,'_space-fsLR-32k-32k_desc-lh_midthickness.surf.gii') )
f32k.mid.rh <- read.fs.surface(filepath = paste0(dir_surf, subjectID,'_space-fsLR-32k-32k_desc-rh_midthickness.surf.gii') )
# Plot the surface
cml = coloredmesh.from.preloaded.data(f32k.mid.lh, morph_data = rep(0, nrow(f32k.mid.lh$vertices)), makecmap_options = list('colFn'=grays) )
cmr = coloredmesh.from.preloaded.data(f32k.mid.rh, morph_data = rep(0, nrow(f32k.mid.rh$vertices)), makecmap_options = list('colFn'=grays) )
brainviews(views = 't4', coloredmeshes=list('lh'=cml, 'rh'=cmr), draw_colorbar = FALSE,
rglactions = list('trans_fun'=limit_fun(-1, 1), 'no_vis'=F))
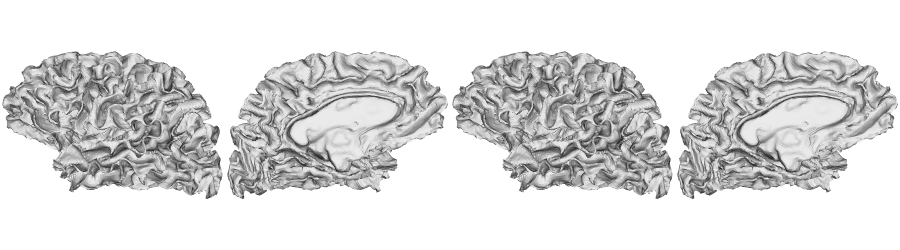
fsLR-32k: White matter surface
# Native fsLR-32k white matter
fsLR32k_wm_lh = read_surface(dir_surf+subjectID+'_hemi-L_space-nativepro_surf-fsLR-32k_label-white.surf.gii', itype='gii')
fsLR32k_wm_rh = read_surface(dir_surf+subjectID+'_hemi-R_space-nativepro_surf-fsLR-32k_label-white.surf.gii', itype='gii')
# Plot the surface
plot_hemispheres(fsLR32k_wm_lh, fsLR32k_wm_lh, size=(900, 250), zoom=1.25, embed_nb=True, interactive=False, share='both',
nan_color=(0, 0, 0, 1), color_range=(1.5, 4), cmap='Greys', transparent_bg=False)
# Set the path to the surface
f32k.wm.lh <- read.fs.surface(filepath = paste0(dir_surf, subjectID,'_space-fsLR-32k-32k_desc-lh_white.surf.gii') )
f32k.wm.rh <- read.fs.surface(filepath = paste0(dir_surf, subjectID,'_space-fsLR-32k-32k_desc-rh_white.surf.gii') )
# Plot the surface
cml = coloredmesh.from.preloaded.data(f32k.wm.lh, morph_data = rep(0, nrow(f32k.wm.lh$vertices)), makecmap_options = list('colFn'=grays) )
cmr = coloredmesh.from.preloaded.data(f32k.wm.rh, morph_data = rep(0, nrow(f32k.wm.rh$vertices)), makecmap_options = list('colFn'=grays) )
brainviews(views = 't4', coloredmeshes=list('lh'=cml, 'rh'=cmr), draw_colorbar = FALSE,
rglactions = list('trans_fun'=limit_fun(-1, 1), 'no_vis'=F))
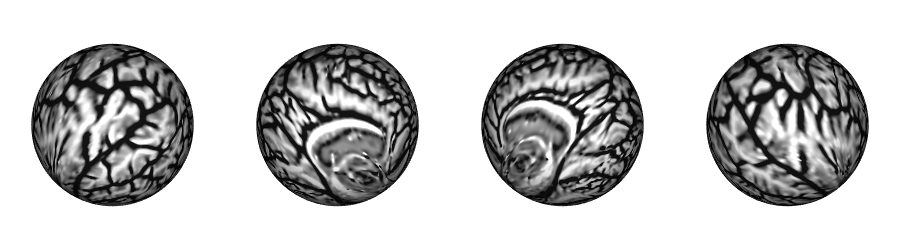
Native sphere
# Native sphere
sph_lh = read_surface(dir_surf+subjectID+'_hemi-L_surf-fsnative_label-sphere.surf.gii', itype='gii')
sph_rh = read_surface(dir_surf+subjectID+'_hemi-R_surf-fsnative_label-sphere.surf.gii', itype='gii')
# Plot the surface
plot_hemispheres(sph_lh, sph_rh, array_name=cv, size=(900, 250), zoom=1.25, embed_nb=True, interactive=False, share='both',
nan_color=(0, 0, 0, 1), color_range=(-0.2, 0.2), cmap="gray", transparent_bg=False)
# Colormap
grays <- colorRampPalette(c('white', 'gray65','black'))
# Set the path to the surface
sph.lh <- read.fs.surface(filepath = paste0(dir_surf, subjectID,'_lh_sphereReg.surf.gii'))
sph.rh <- read.fs.surface(filepath = paste0(dir_surf, subjectID,'_rh_sphereReg.surf.gii'))
# Set the color limits
lf= limit_fun(-0.2, 0.2)
# Create the coloredmeshes
cml = coloredmesh.from.preloaded.data(sph.lh, morph_data = lf(read.fs.mgh(cv.lh)), hemi = 'lh', makecmap_options = list('colFn'=grays))
cmr = coloredmesh.from.preloaded.data(sph.rh, morph_data = lf(read.fs.mgh(cv.rh)), hemi = 'rh', makecmap_options = list('colFn'=grays))
sph.nat <- brainviews(views = 't4', coloredmeshes=list('lh'=cml, 'rh'=cmr), rglactions = list('no_vis'=T))
# Plot the surface
plot_surface(sph.nat, 'Native sphere curvature [1/mm]')
Superficial White Matter (SWM) in fsnative surface
The superficial white matter surfaces are generated across 3 different surface layer from the white mater to 1, 2 and 3mm deeps.
Then each quantitative map from /maps is resample from fsnative to fsaverage5, fsLR-32k and fsLR-5k. In this example we will only plot the native surfaces.
SWM Surfaces
# Function to load and plot each SWM surfaces
def plot_swm(mm='1'):
# SWM fsnative 1mm
swm_lh = read_surface(f'{dir_surf}{subjectID}_hemi-L_surf-fsnative_label-swm{mm}.0mm.surf.gii', itype='gii')
swm_rh = read_surface(f'{dir_surf}{subjectID}_hemi-R_surf-fsnative_label-swm{mm}.0mm.surf.gii', itype='gii')
# Plot the surface
fig = plot_hemispheres(swm_lh, swm_rh, size=(900, 250), zoom=1.25, embed_nb=True, interactive=False, share='both',
nan_color=(0, 0, 0, 1), color_range=(1.5, 4), cmap='Greys', transparent_bg=False)
return(fig)
# SWM 1,2,3mm
for (mm in 1:3) {
# Set the path to the surface
f32k.swm.lh <- read.fs.surface(filepath = paste0(dir_surf, subjectID,'_hemi-L_surf-fsnative_label-swm',mm,'.0mm.surf.gii') )
f32k.swm.rh <- read.fs.surface(filepath = paste0(dir_surf, subjectID,'_hemi-R_surf-fsnative_label-swm',mm,'.0mm.surf.gii') )
# Plot the surface
cml = coloredmesh.from.preloaded.data(f32k.swm.lh, morph_data = rep(0, nrow(f32k.swm.lh$vertices)), makecmap_options = list('colFn'=grays) )
cmr = coloredmesh.from.preloaded.data(f32k.swm.rh, morph_data = rep(0, nrow(f32k.swm.rh$vertices)), makecmap_options = list('colFn'=grays) )
brainviews(views = 't4', coloredmeshes=list('lh'=cml, 'rh'=cmr), draw_colorbar = FALSE,
rglactions = list('trans_fun'=limit_fun(-1, 1), 'no_vis'=F))
}
/maps: fsnative, fsaverage5, fsLR-32k and fsLR-5k
Each file map with the extension
func.giicorresponds to the data map from a NIFTI image at a certain deep.The deep from where it was mapped is in the name after the string
label-.The hemisphere is either
Lfor left orRfor right.The surface will match the number of points of the surface that corresponds that file map. The options are:
fsnative,fsLR-32k,fsLR-5kandfsaverage5.
Note ❕
For example the file below corresponds to the left native surface mapped from midthicknes of the T1map nifti image:
sub-001_hemi-L_surf-fsnative_label-midthickness_T1map.func.gii
The maps on the surfaces
fsnative,fsLR-32k,fsLR-5k, can be plot on their native surface or on the standard surface (regular or inflated).
Warning
There is NO inherent smoothing applied to the map. If the user desires smoothing, they should customize it according to their preferences and requirements.
def load_qmri(qmri='', surf='fsLR-32k'):
'''
This function loads the qMRI intensity maps from midthickness surface
'''
# List the files
files_lh = sorted(glob.glob(f"{dir_maps}/*_hemi-L_surf-{surf}_label-midthickness_{qmri}.func.gii"))
files_rh = sorted(glob.glob(f"{dir_maps}/*_hemi-R_surf-{surf}_label-midthickness_{qmri}.func.gii"))
# Load map data
surf_map=np.concatenate((nib.load(files_lh[0]).darrays[0].data, nib.load(files_rh[0]).darrays[0].data), axis=0)
return(surf_map)
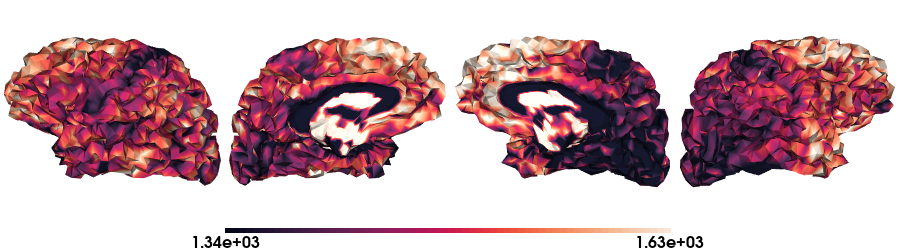
def plot_qmri(qmri='', surf='fsLR-32k', label='pial', cmap='rocket', rq=(0.15, 0.95)):
'''
This function plots the qMRI intensity maps on the pial surface
'''
# Load the data
map_surf = load_qmri(qmri, surf)
print('Number of vertices: ' + str(map_surf.shape[0]))
# Load the surfaces
surf_lh=read_surface(f'{dir_surf}/{subjectID}_hemi-L_space-nativepro_surf-{surf}_label-{label}.surf.gii', itype='gii')
surf_rh=read_surface(f'{dir_surf}/{subjectID}_hemi-R_space-nativepro_surf-{surf}_label-{label}.surf.gii', itype='gii')
# Color range based in the quantiles
crange=(np.quantile(map_surf, rq[0]), np.quantile(map_surf, rq[1]))
# Plot the group T1map intensitites
fig = plot_hemispheres(surf_lh, surf_rh, array_name=map_surf, size=(900, 250), color_bar='bottom', zoom=1.25, embed_nb=True, interactive=False, share='both',
nan_color=(0, 0, 0, 1), cmap=cmap, color_range=crange, transparent_bg=False, screenshot = False)
return(fig)
# Under construction
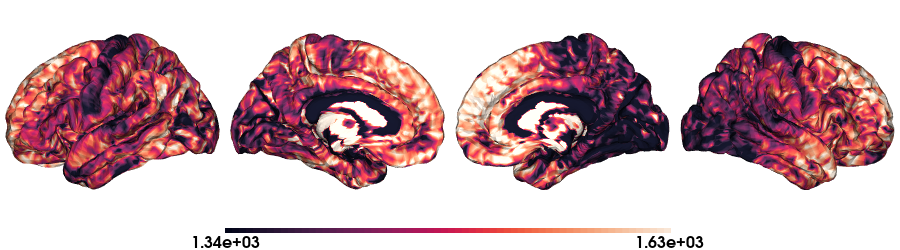
/maps: fsaverage5, fsLR-32k and fsLR-5k on standard
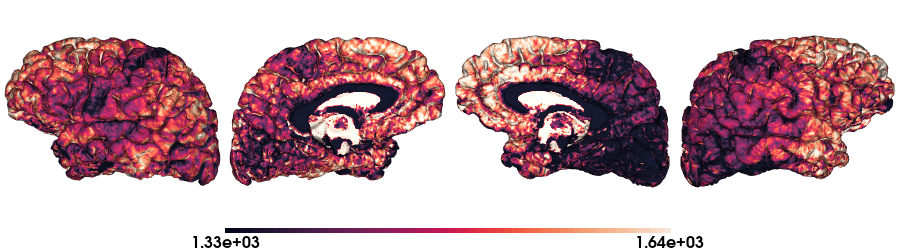
T1map on fsaverage5 stardard
# Load the T1map data on fsaverage5
map_data = load_qmri('T1map', 'fsaverage5')
# Color range based in the quantiles
crange=(np.quantile(map_data, 0.15), np.quantile(map_data, 0.95))
# Plot data on standard surface
plot_hemispheres(fs5_lh, fs5_rh, array_name=map_data, size=(900, 250), color_bar='bottom', zoom=1.25, embed_nb=True, interactive=False, share='both',
nan_color=(0, 0, 0, 1), color_range=crange, cmap="rocket", transparent_bg=False)
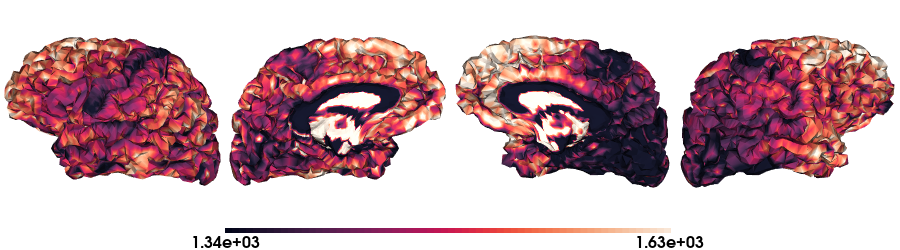
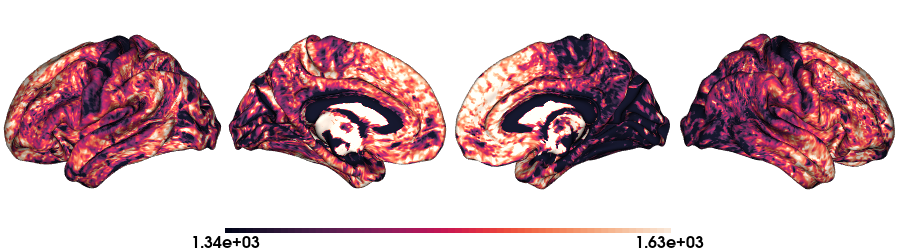
T1map on fsLR-32k stardard
# Load the T1map data on fsLR-32k
map_data = load_qmri('T1map', 'fsLR-32k')
# Plot data on standard surface
plot_hemispheres(f32k_lh, f32k_rh, array_name=map_data, size=(900, 250), color_bar='bottom', zoom=1.25, embed_nb=True, interactive=False, share='both',
nan_color=(0, 0, 0, 1), color_range=crange, cmap="rocket", transparent_bg=False)
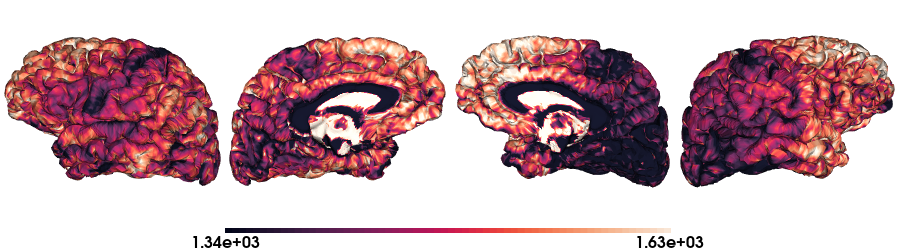
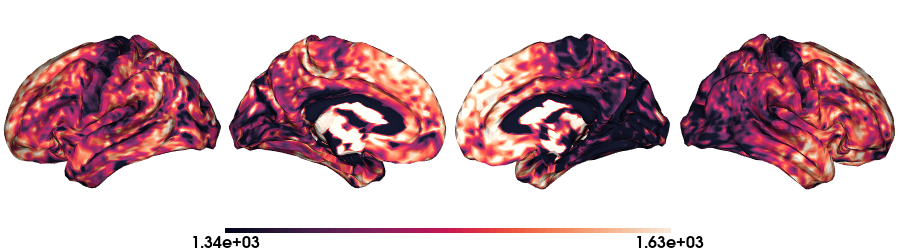
T1map on fsLR-5k stardard
# Load the T1map data on fsLR-5k
map_data = load_qmri('T1map', 'fsLR-5k')
# Plot data on standard surface
plot_hemispheres(f5k_lh, f5k_rh, array_name=map_data, size=(900, 250), color_bar='bottom', zoom=1.25, embed_nb=True, interactive=False, share='both',
nan_color=(0, 0, 0, 1), color_range=crange, cmap="rocket", transparent_bg=False)
Atlas labels on surface
All the native surface labels generated by micapipe are stored inside the subject’s freesurfer directory.
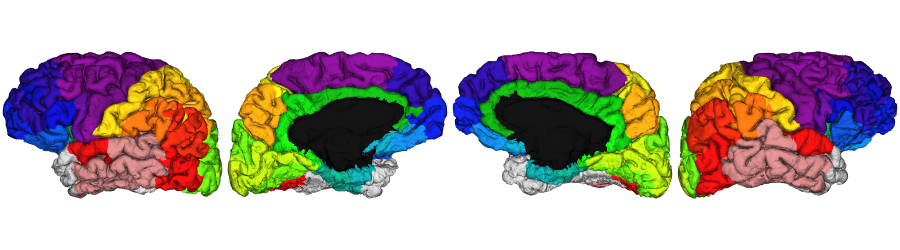
Schaefer-400 labels
# Load annotation file
annot = 'schaefer-400'
annot_lh= dir_FS + '/label/lh.' + annot + '_mics.annot'
annot_rh= dir_FS + '/label/rh.' + annot + '_mics.annot'
label = np.concatenate((nib.freesurfer.read_annot(annot_lh)[0], nib.freesurfer.read_annot(annot_rh)[0]), axis=0)
# plot labels on surface
plot_hemispheres(pial_lh, pial_rh, array_name=label, size=(900, 250), zoom=1.25, embed_nb=True, interactive=False, share='both',
nan_color=(0, 0, 0, 1), cmap='nipy_spectral', transparent_bg=False)
# Plot the surface
schaefer.400 <- vis.subject.annot('freesurfer/', subjectID, 'schaefer-400_mics', 'both', surface='pial',
views=NULL, rglactions = list('no_vis'=T))
plot_surface(schaefer.400, 'Schaefer-400')
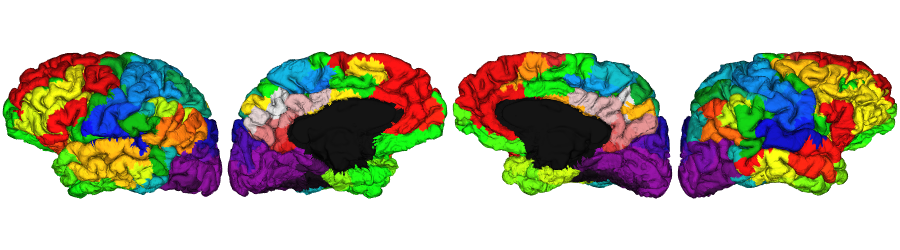
Extra: Economo labels
# Load annotation file
annot = 'economo'
annot_lh= dir_FS + '/label/lh.' + annot + '_mics.annot'
annot_rh= dir_FS + '/label/rh.' + annot + '_mics.annot'
label = np.concatenate((nib.freesurfer.read_annot(annot_lh)[0], nib.freesurfer.read_annot(annot_rh)[0]), axis=0)
# plot labels on surface
plot_hemispheres(pial_lh, pial_rh, array_name=label, size=(900, 250), zoom=1.25, embed_nb=True, interactive=False, share='both',
nan_color=(0, 0, 0, 1), cmap='nipy_spectral', transparent_bg=False)
# Plot the surface
economo <- vis.subject.annot('freesurfer/', subjectID, 'economo_mics', 'both', surface='pial',
views=NULL, rglactions = list('no_vis'=T))
plot_surface(economo, 'economo', img_only=TRUE)