Microstructural profile covariance
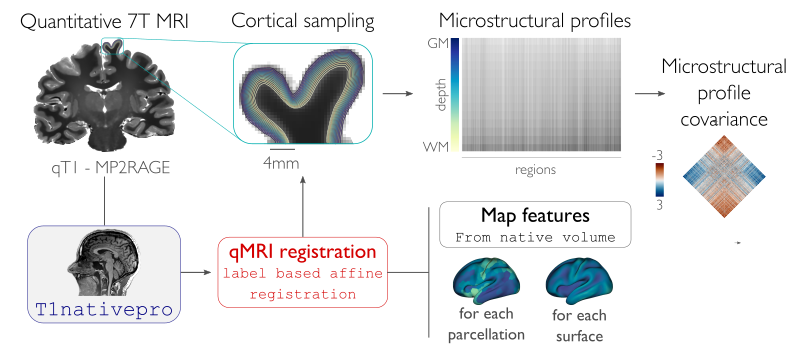
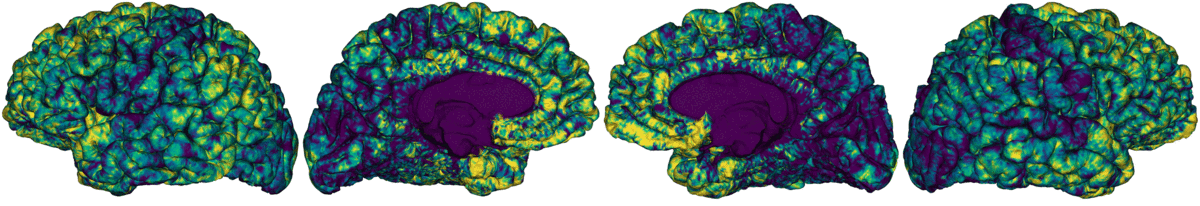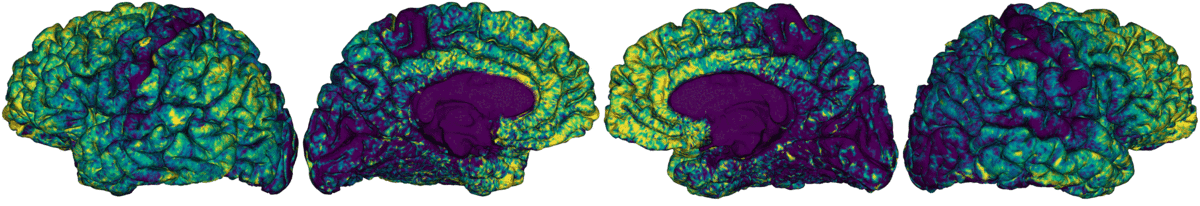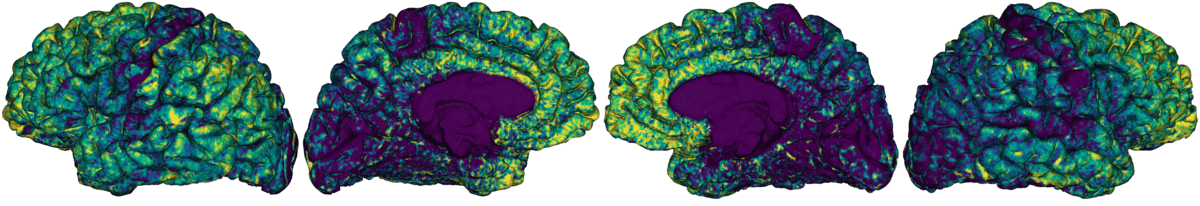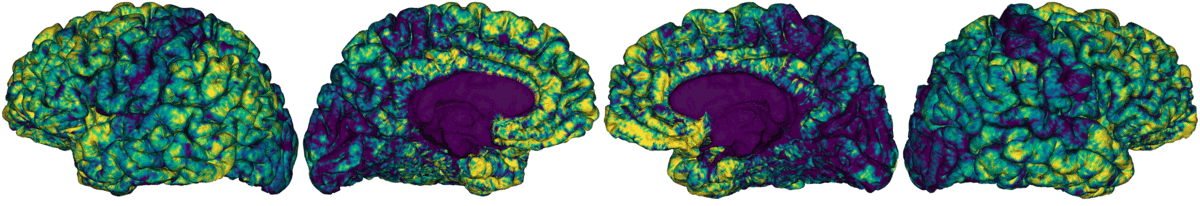
This module samples intracortical intensities from a microstructurally-sensitive contrast. This is achieved by constructing a series of equivolumetric surfaces between pial and white matter boundaries, yielding unique intracortical intensity profiles at each vertex of the native surface mesh. This approach has been previously applied in the whole cortex, as well as in targeted structures such as the insula. By parcellating and cross-correlating nodal intensity profiles, this module profiles microstructural profile covariance (MPC) matrices describing similiarity in intracortical microstructure across the cortex.
Intracortical equivolumetric surfaces are generated using scripts from the surface tools repository, available via GitHub.
-MPC
Prerequisites 🖐🏼
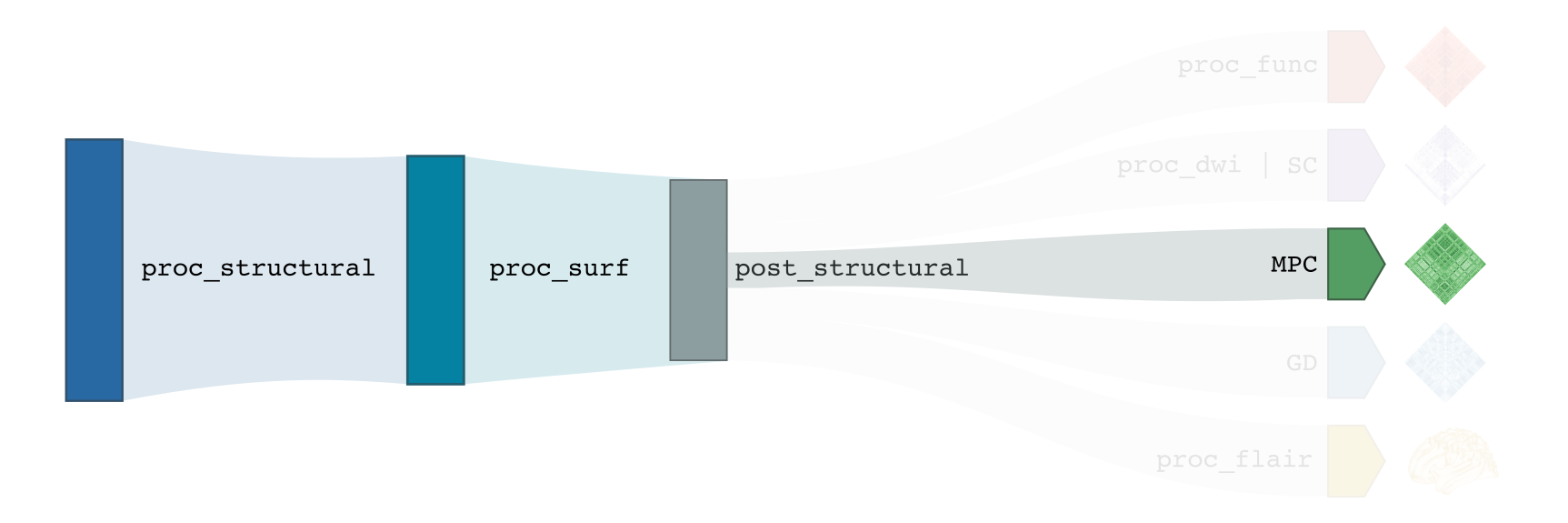
You need to run -proc_structural, -proc_surf and -post_structural before this stage
Compute registration from qMRI to surface native space.
Generate 16 equivolumetric surfaces between pial and white matter boundary. Surfaces closest to pial and white matter boundaries are then discarded to account for partial volume effects, resulting in 14 surfaces used for further analyses
Perform surface-based registration to fsaverage5, fsLR-32k, fsLR-5k templates
Average intensity profiles within parcels defined on the native surface, while excluding outlier vertices
Perform partial correlation, controlling for parcel-wide mean profile, across all pairs of intensity profiles
Compute registration from qMRI to T1w-nativepro space.
Map qMRI midthickness and white surfaces to fsLR-32k, fsLR-5k and fsaverage5
Terminal:
$ micapipe -sub <subject_id> -out <outputDirectory> -bids <BIDS-directory> -MPC <options>
Optional arguments:
-MPC has several optional arguments:
Optional argument |
Description |
|---|---|
|
Specifies an input quantitative MRI on which to sample intensities for the MPC analysis. By default the pipeline will search this regex anat/*mp2rage*T1map.nii* You must specify this flag with the full path to your qMRI (for example, MTR, MTSAT, T2star, T1w/T2w). |
|
Path to scan which will be register to the surface ( e.g INV1, better anatomical contrast ). It MUST be on the same space as the main microstructural image!!.If it is EMPTY will try to find a T1map from here: anat/mp2rage*T1map.nii. Set to ‘FALSE’ to use microstructural_img for registrations. |
|
Default=qMRI. Provide a string with this this flag to process new quantitative map. ( this will create a new directory here: anat/surf/micro_profiles/acq-<mpc_acq> ) |
|
Specify this option to perform the registration based on synthseg |
|
Specify this option to perform the NON-Linear registration (e.g. MRI with strong geometric distortions). |
Directories created or populated by -MPC:
- <outputDirectory>/micapipe_v0.2.0/<sub>/mpc/<mpc_acq>
- <outputDirectory>/micapipe_v0.2.0/<sub>/anat
- <outputDirectory>/micapipe_v0.2.0/<sub>/xfm
Files generated by -MPC:
- Microstructural image in native Surfer space:
<outputDirectory>/micapipe_v0.2.0/<sub>/anat/<sub>_space-fsnative_<mpc_acq>.nii.gz
<outputDirectory>/micapipe_v0.2.0/<sub>/anat/<sub>_space-fsnative_<mpc_acq>.nii.gz
- Boundary-based registration outputs from microstructural image to native FreeSurfer space:
<outputDirectory>/micapipe_v0.2.0/<sub>/xfm/
<sub>_from-T1map_to-fsnative_0GenericAffine.mat
<sub>_from-T1map_to-nativepro_0GenericAffine.mat
<sub>_from-T1map_to-fsnative0_1Warp.nii.gz (optional)
<sub>_from-T1map_to-nativepro_1Warp.nii.gz (optional)
<sub>_from-T1map_to-fsnative0_1InverseWarp.nii.gz (optional)
<sub>_from-T1map_to-nativepro_1InverseWarp.nii.gz (optional)
- Equivolumetric surface sampling ouputs stored in <outputDirectory>/micapipe_v0.2.0/mpc/<mpc_acq>:
- MPC json card:
<sub>_MPC-<mpc_acq>.json
- Native intensity profiles:
<sub>_surf-fsnative_desc-intensity_profiles.shape.gii
- fsaverage5 intensity profiles:
<sub>_surf-fsaverage5_desc-intensity_profiles.shape.gii
- fsLR-32k intensity profiles:
<sub>_surf-fsLR-32k_desc-intensity_profiles.shape.gii
- fsLR-5k intensity profiles and MPC:
<sub>_surf-fsLR-5k_desc-intensity_profiles.shape.gii
<sub>_surf-fsLR-5k_MPC.shape.gii
- Parcellated intensity profiles:
<sub>_atlas-<atlas>_desc-intensity_profiles.shape.gii
- MPC matrices:
<sub>_atlas-<atlas>_desc-MPC.shape.gii